 AI
AI
 AI
AI
 AI
AI
Alphabet Inc.’s artificial intelligence research lab DeepMind Technologies said today it has solved a 50-year-old “grand challenge” in biology by creating software that can predict the atomic structure that proteins will fold into in a matter of days.
The breakthrough could pave the way for a better understanding of diseases and new drug discoveries.
DeepMind’s software, known as “AlphaFold,” has solved what’s known as the “protein folding problem,” which refers to attempts to understand how a protein’s amino acid sequence shapes its 3D atomic structure. The form it folds into is dictated by interatomic forces and thermodynamics, and it’s extremely difficult to predict.
It’s an important discovery because every living cell contains thousands of different proteins that keep it alive and functioning. By predicting the shape a protein folds into, scientists can determine its function, and nearly all diseases, including cancer and dementia, are related to how proteins function.
The American biologist Christian Boehmer Anfinsen Jr. won the 1972 Nobel Prize in chemistry for theorizing that a protein’s amino acid sequence should fully determine its structure, and that hypothesis sparked a five-decade search for a computational simulation that could predict a protein’s structure based on its 1D amino acid sequence.
“A major challenge, however, is that the number of ways a protein could theoretically fold before settling into its final 3D structure is astronomical,” DeepMind’s AlphaFold team wrote in a blog post. “In 1969 Cyrus Levinthal noted that it would take longer than the age of the known universe to enumerate all possible configurations of a typical protein by brute force calculation – Levinthal estimated 10^300 possible conformations for a typical protein.”
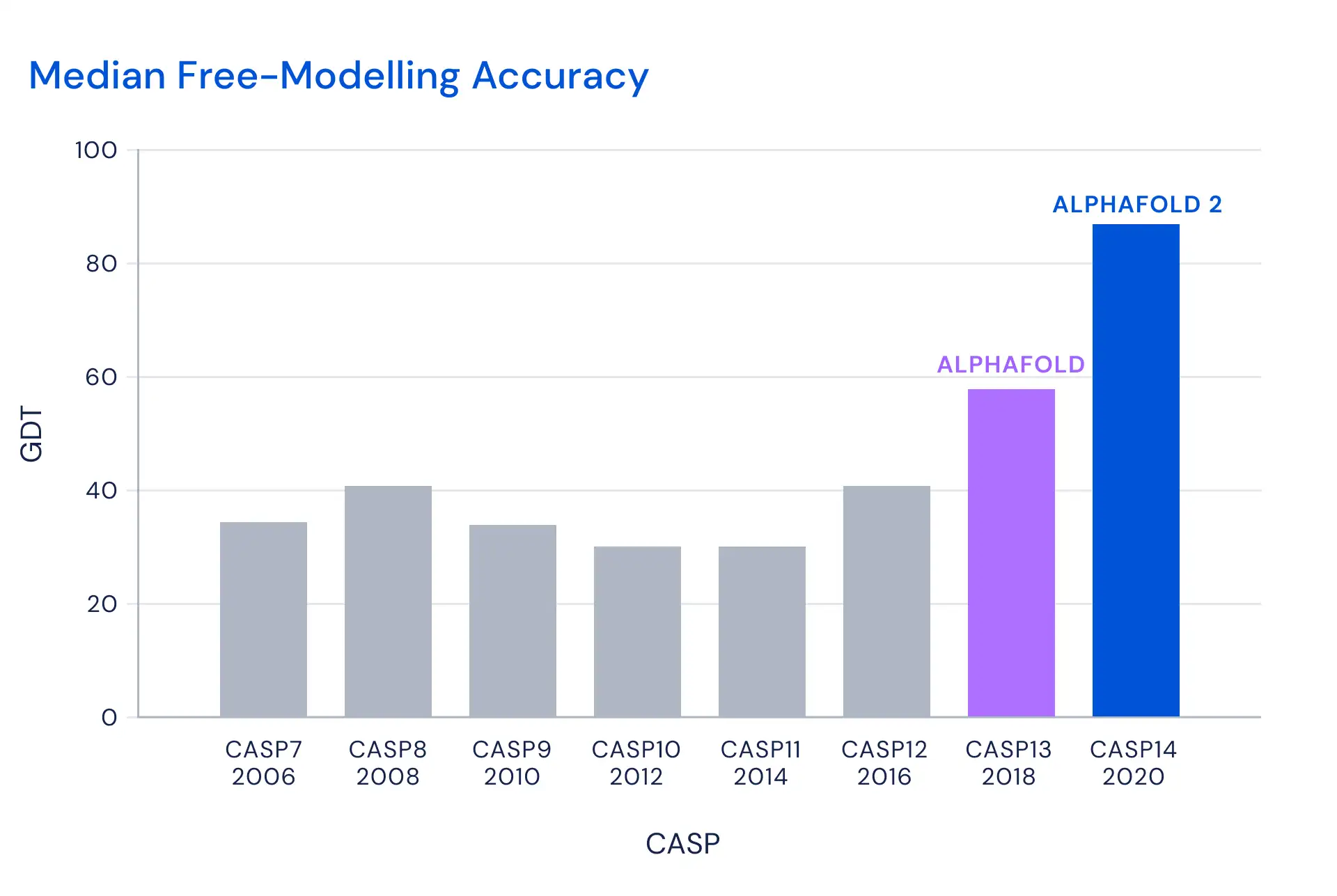
The Critical Assessment for Structure Prediction, or CASP, was established in 1994 as a biannual challenge that aimed to catalyze research into the problem. CASP’s measure of success is the Global Distance Test, which is based on the percentage of amino acid residues that are predicted within a threshold distance of their correct position. The possible scores range from 0-100, with the unofficial benchmark being anything above 90 GDT.
DeepMind said finally cracked the problem in its 14th attempt, CASP14, scoring 92.4 GDT, which means its predictions have an average error of just 1.6 Angstroms, comparable to the width of an atom, or just 0.1 nanometer. That’s a significant improvement over its last effort, in 2018, which scored less than 60 GDT.
“For the latest version of AlphaFold, used at CASP14, we created an attention-based neural network system, trained end-to-end, that attempts to interpret the structure of this graph, while reasoning over the implicit graph that it’s building,” DeepMind explained. “It uses evolutionarily related sequences, multiple sequence alignment (MSA), and a representation of amino acid residue pairs to refine this graph.”
“We have been stuck on this one problem – how do proteins fold up – for nearly 50 years,” said Professor John Moult, CASP chair and co-founder. “To see DeepMind produce a solution for this, having worked personally on this problem for so long and after so many stops and starts, wondering if we’d ever get there, is a very special moment.”
DeepMind said the AlphaFold model was created using a neural network that runs on 128 of Google LLC’s latest TPU neural processing cores, crunching data from 170,000 protein from public and other databases. It took “a few weeks” to train the model, which is able to successfully predict the final structure of new proteins in just a few days time.
Analyst Charles King of Pund-IT Inc. told SiliconANGLE that DeepMind has scored an impressive achievement that could fundamentally alter the way scientists and clinicians understand and address protein structures.
“The DeepMind team and Alphabet should be over the moon with surpassing this milestone,” King said. “While solving the protein folding challenge certainly highlights the potential and potential value of AI, it also underscores the continuing vital role that computational technologies have in modern scientific inquiry. This is the latest peak but it was attained following past structural achievements that were achieved, brick by brick, through the efforts of countless scientists and technologists.”
“Not surprisingly the problem was cracked using a neural network,” said Constellation Research Inc. analyst Holger Mueller. “It’s another achievement that underlines Google’s leading position in AI.”
Next, DeepMind will attempt to make the AlphaFold system accessible in a scalable way so that third-party researchers can better understand how different proteins impact specific diseases and try to create new treatments for them.
Support our mission to keep content open and free by engaging with theCUBE community. Join theCUBE’s Alumni Trust Network, where technology leaders connect, share intelligence and create opportunities.
Founded by tech visionaries John Furrier and Dave Vellante, SiliconANGLE Media has built a dynamic ecosystem of industry-leading digital media brands that reach 15+ million elite tech professionals. Our new proprietary theCUBE AI Video Cloud is breaking ground in audience interaction, leveraging theCUBEai.com neural network to help technology companies make data-driven decisions and stay at the forefront of industry conversations.